What does PLIER do?
PLIER is a matrix decomposition method that summarizes gene expression as a product of a small set of latent variables (matrix B) and gene loadings (matrix Z).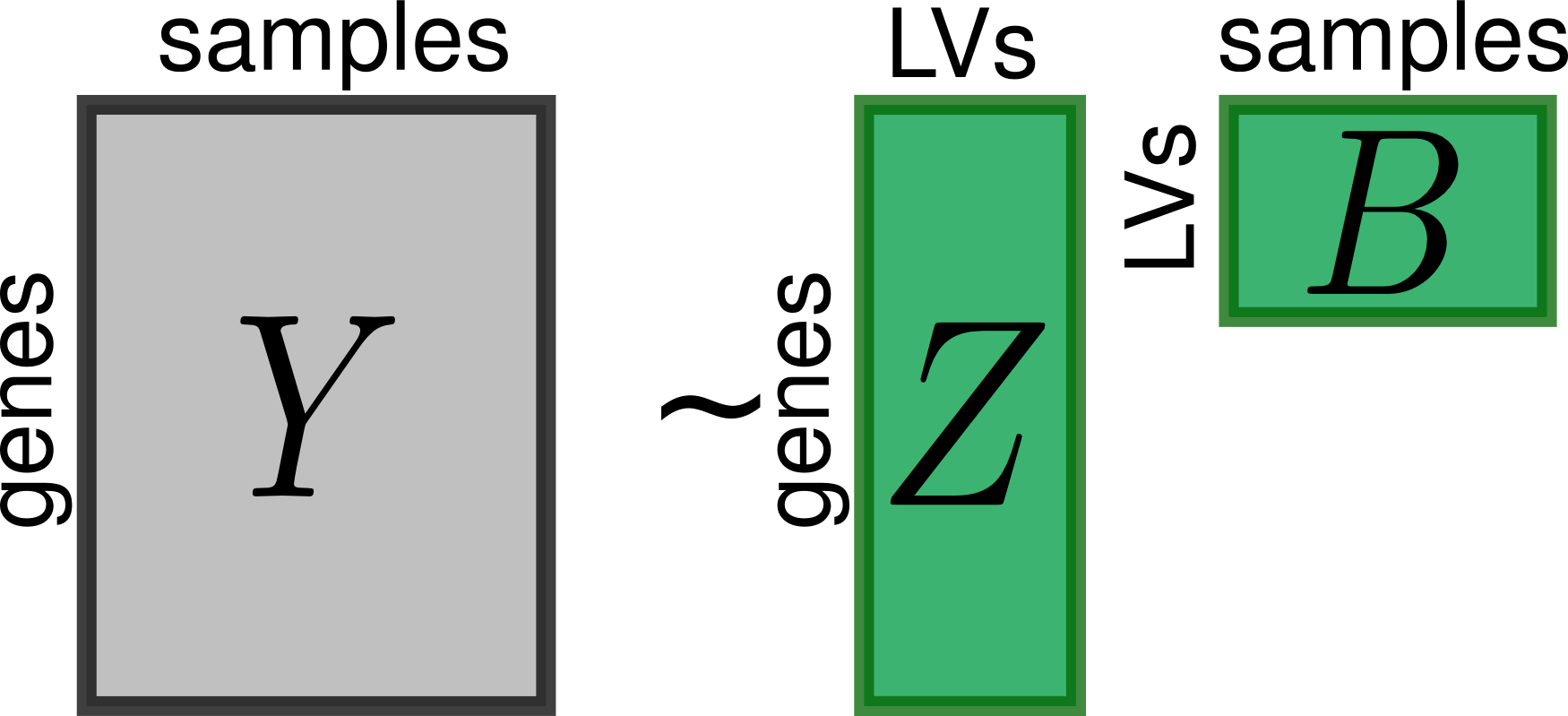
It also enforces a correspondence between the loadings (matrix Z) and some sparse combinations of a prior information genesets (matrix C).
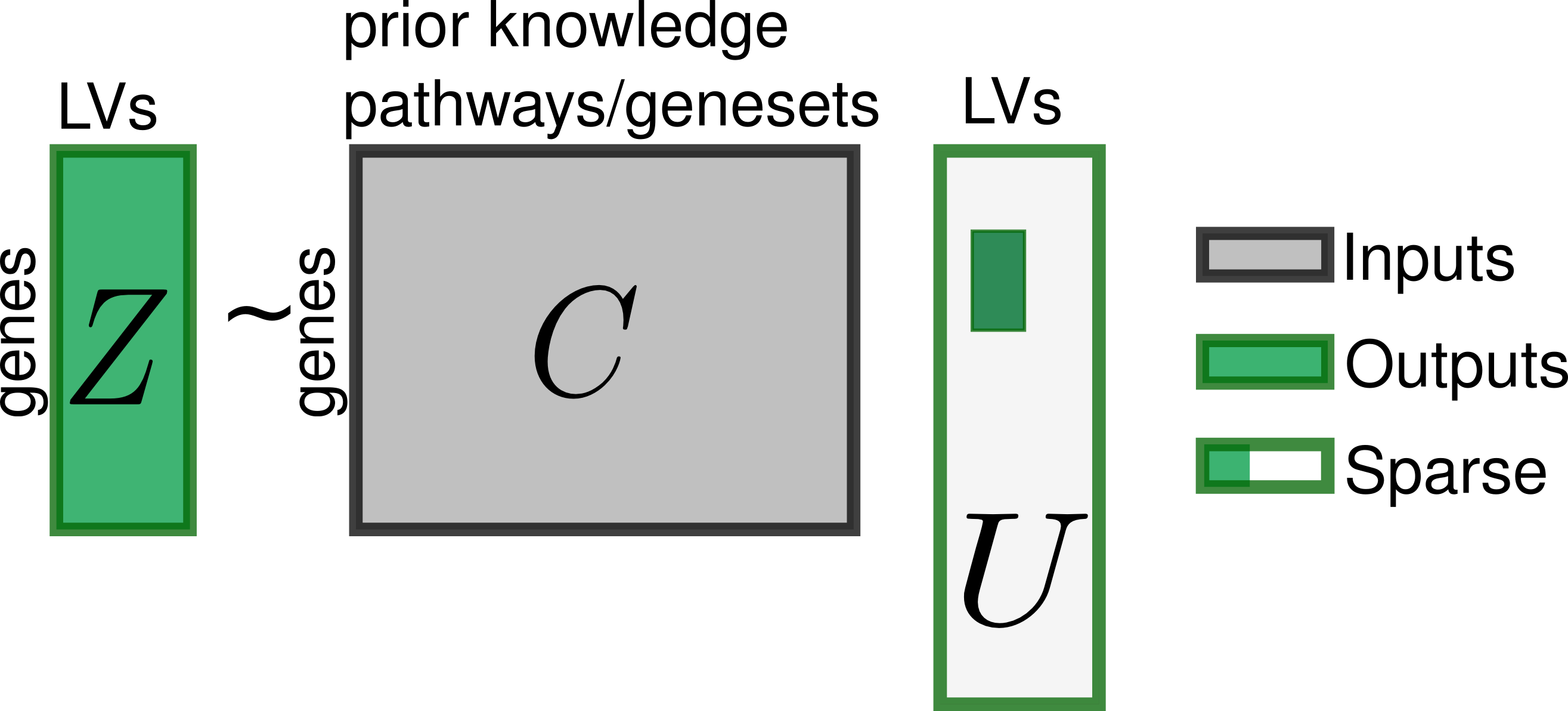
The inferred latent variables can be directly connected to a specific biological cause such as cell-type composition or pathway activity. PLIER will even name the latent variables in matrix B with its best guess of what they represent.
What is the point?
The latent variables contained in matrix B give a summary view of the dataset and can be plugged into the same statistical pipelines that are used to analyze gene-level measurements. The latent representation reduces redundancy, since correlated genes are summarized, and provides noise reduction, as all latent variables are automatically corrected for all other sources of systematic variation.Moreover, since many of the latent variables can be easily interpreted as providing estimates of composition or pathway activity, downstream statistical analysis can provide biologically tangible hypotheses. For example: patient group A has higher type-II interferon signalling than patient group B.
How is this different from pathway-level analysis of differential expression results?
There are several important distinctions:- Only the pathways for which there is support in the data are considered.
- Redundant pathways, ones that share many genes or whose expression patterns cannot be separated, are collapsed.
- The matrix decomposition framework ensures that distinct pathway-level effects are maximally isolated from others.
- PLIER provides pathway-level estimates independently of the downstream statistical analysis which means that the same estimates can be reused to test many models and hypotheses.